As a prospective scientist one of my goals for this year was to narrow my field of view for the future! I have always had a fascination with genomics and the fact that any one of our cells can tell us so much information about our surroundings and ourselves. I am interested in finding an intersection between genetic techniques and conservation applications. For this reason, I decided to spend a week with @Lab in La Paz, BCS, Mexico.
This Lab is was started by Adrian Munguia Vega, an accomplished marine geneticist who works here in La Paz, and also at the University of Arizona in the United States. Adrian aims to use genetic tools for management and conservation purposes in any model system around him. Totally up my alley! He is currently working on a huge undertaking to understand the Marine Biodiversity in the Mexican Pacific and in the Gulf of California using eDNA.

First a little background: I LOVE eDNA! I think it is the coolest thing, and I love learning about its applications.
eDNA stands for environmental DNA, and it is exactly that – DNA that can be extracted from any environmental sample, such as water, soil or feces, without first targeting an organism.

As a marine scientist, this is particularly exciting as it implies that just by collecting a water sample we can identify what is around. This is done by pumping a bunch of water through a tiny paper filter that concentrates the entire volume of DNA present in the sample. We then can extract this DNA, and use it to address whatever questions we are interested in answering.

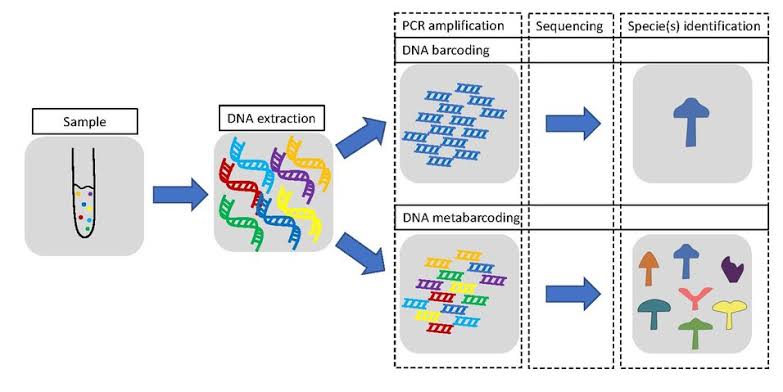
For diversity that is difficult to detect using visual methods, or hard to sample, environmental DNA (eDNA) provides a cost effective, and more efficient method to find the invisible. Currently the most common use of eDNA is to quantify biodiversity, or the number and identity of species present in an area, using a method called DNA Barcoding. Once the DNA is extracted we can amplify a DNA sequence of interest or “barcode”, that can then be used to identify a species from an existing database of these sequences. Just like scanning the barcode of a box of cookies at a grocery store will tell the clerk what type of cookies they are, the DNA barcodes of different species can tell the biologist what type of organism they are! These “barcode” gene sequences are chosen based off of 3 requirements.
- The gene should be highly conserved and thus present in the majority of organisms – this allows us to make the assumption that unknown species will have this gene. Because of this requirement we often use genes that are necessary for survival, and thus very likely to be present
- Each cell contains many copies of the gene – this implies that when sample quantity is limited like in eDNA, you are more likely to have copies of your gene of interest
- The gene should be the same within species, but different enough to identify different species, but also similar enough that a non-targeted primer can amplify the gene – this is a bit confusing. I shall break it down for you
- To amplify these sequences from a DNA sample we use primers; these are little codes that can find and stick to both ends of our sequence. For these primers to work on most organisms, the gene has to be conserved enough that the same primer can find and stick to the sequence in any organism
- Since we are using the sequence to identify species, we want it to have enough differences so it can act as a barcode that can tell the difference between lets say a chocolate chip cookie, and an oatmeal one
- However we don’t want it to be so different that it will tell the difference between a chocolate chip cookie and a chocolate chunk cookie (the same species in this argument)
Once a good target gene is chosen, it can be amplified and used to identify what species are present in a sample of Water. This is all well and done in practice, but it requires a lot of existing knowledge to function. Like the barcode of a cookie box has a database telling the clerk it is chocolate chip, for meta barcoding (barcoding of many organisms at once), we need a database that can tell us what is what in a sample of eDNA.
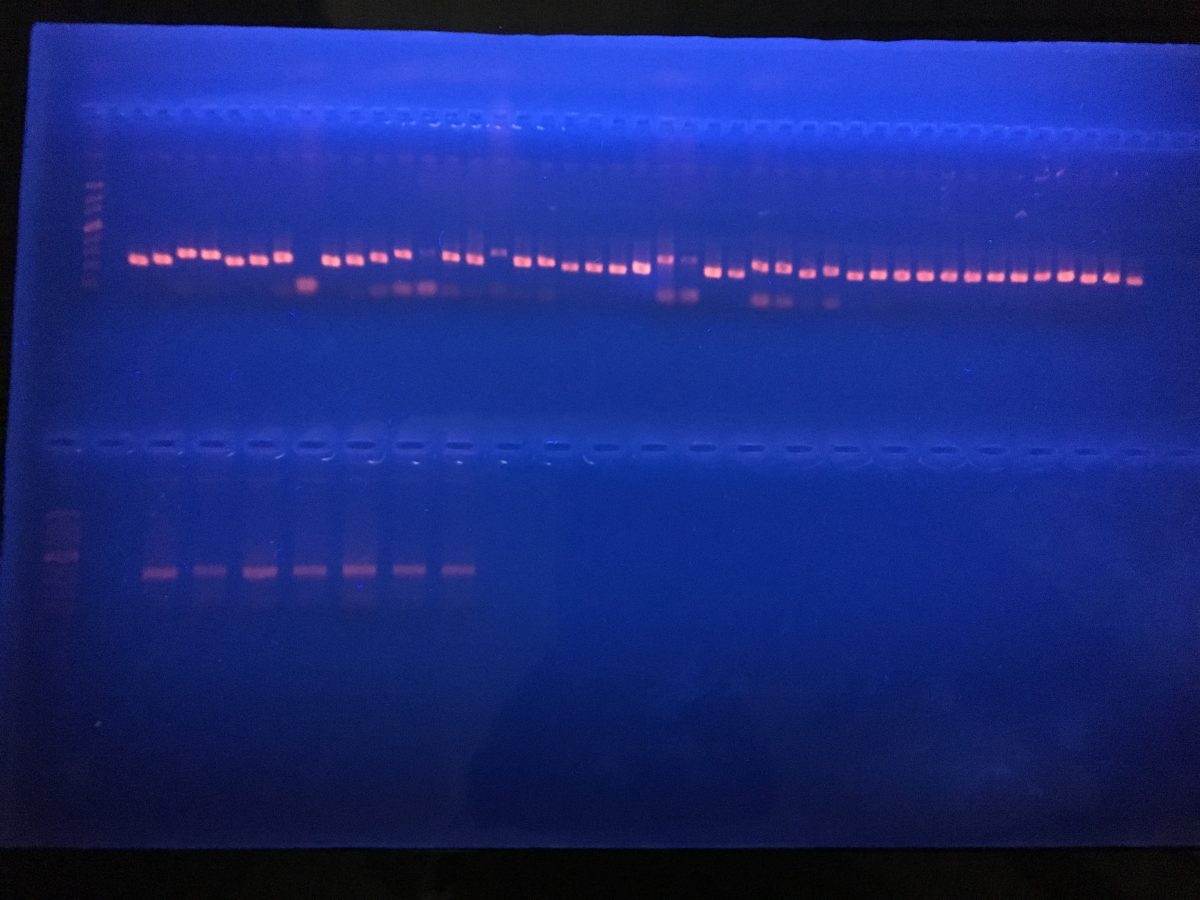
This brings me to the little project I have been working on while I am here. With his students and other scientists, Adrian is working on a project with Cryptic Fish. They play a vital role in the ecosystem, as the nutritional link between zooplankton and conspicuous reef fish. Because of their inaccessibility as tiny fish hiding away, they have been historically understudied. Cami MacLoughlin, a PhD candidate at @Lab is studying their presence and distribution in the Gulf of California. These organisms are perfect candidates for eDNA studies, because as their name suggests they are difficult to identify and monitor. Over three intensive field expeditions, and 280 hours underwater, they collected over 2000 specimens, over 17 families and 80 species. Using these samples I am starting to build a reference library that can be used in the future as barcodes! Once the sequence of interest is amplified it will be sequenced and then uploaded to a database. Next time anybody is studying any of these species, they will already be logged and ready to find!
Though the type of lab work I am doing here I have done before, I am learning a lot from the people, the new species and new concepts! DNA Barcoding and eDNA are relatively new fields, and their applications are growing. I am hoping to work in these fields in the future, so this experience has been very valuable to me. I am really loving La Paz; it is a hub of marine science in Mexico!
Thank you to Rolex and Our World-Underwater Scholarship Society https://www.owuscholarship.org/ for allowing me to work with such incredible people!